Volume 6, Issue 3 (2025)
J Clinic Care Skill 2025, 6(3): 147-154 |
Back to browse issues page
Article Type:
Subject:
History
Received: 2025/05/23 | Accepted: 2025/07/1 | Published: 2025/07/5
Received: 2025/05/23 | Accepted: 2025/07/1 | Published: 2025/07/5
How to cite this article
Nazar E, Abdollahi A, Afarinesh Khaki P, Norouzi Shadehi M, Beighmohammadi M, Nateghi S, et al . Antibiotic Susceptibility of Body Fluids and Urine Cultures from Intensive Care Unit Patients in a Tertiary Care Center in Iran. J Clinic Care Skill 2025; 6 (3) :147-154
URL: http://jccs.yums.ac.ir/article-1-424-en.html
URL: http://jccs.yums.ac.ir/article-1-424-en.html
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



Rights and permissions
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Authors
E. Nazar1 
 , A. Abdollahi2
, A. Abdollahi2 
 , P. Afarinesh Khaki3
, P. Afarinesh Khaki3 
 , M. Norouzi Shadehi3
, M. Norouzi Shadehi3 
 , M.T. Beighmohammadi4
, M.T. Beighmohammadi4 
 , S. Nateghi5
, S. Nateghi5 
 , Z. Panahi6
, Z. Panahi6 
 , A. Nozheh *1
, A. Nozheh *1 


 , A. Abdollahi2
, A. Abdollahi2 
 , P. Afarinesh Khaki3
, P. Afarinesh Khaki3 
 , M. Norouzi Shadehi3
, M. Norouzi Shadehi3 
 , M.T. Beighmohammadi4
, M.T. Beighmohammadi4 
 , S. Nateghi5
, S. Nateghi5 
 , Z. Panahi6
, Z. Panahi6 
 , A. Nozheh *1
, A. Nozheh *1 

1- “Department of Pathology, Faculty of Medicine” and “Sina Hospital”, Tehran University of Medical Sciences, Tehran, Iran
2- Department of Pathology, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
3- Central laboratory of Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
4- Department of Intensive Care Medicine, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
5- Department of Cardiology, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
6- Department of Obstetrics and Gynecology, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
2- Department of Pathology, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
3- Central laboratory of Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
4- Department of Intensive Care Medicine, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
5- Department of Cardiology, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
6- Department of Obstetrics and Gynecology, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran, Iran
Full-Text (HTML) (163 Views)
Introduction
Antibiotic resistance has become one of the most important determinants of outcomes in patients with severe infections. A recent estimate reports that more than ten million patients can die each year globally from antibiotic resistance. This rising number reflects a growing public health crisis that threatens to undermine the effectiveness of existing antibiotic treatments. The report emphasizes that global action is necessary to tackle this issue efficiently and sustainably. The impact of such a response is particularly relevant in settings with high rates of multidrug resistance, such as hospitals and, more importantly, intensive care units (ICUs). These critical care units are especially vulnerable due to factors like the frequent use of broad-spectrum antibiotics, prolonged patient stays, invasive procedures, and patients’ compromised immune statuses. In these environments, antibiotic-resistant bacteria can spread rapidly, complicate treatment regimens, and significantly increase mortality rates. Without effective interventions, including enhanced surveillance, antibiotic stewardship programs, and infection prevention and control measures, the burden of antibiotic resistance in ICUs will continue to rise, threatening patient outcomes and healthcare systems worldwide [1-4].
National action plans and surveillance are indeed necessary to mitigate this issue. The problem of antibiotic resistance is an emerging and serious challenge worldwide. Unfortunately, Iran still lacks a well-organized national surveillance system to comprehensively collect and monitor data on antimicrobial resistance and antibiotic susceptibility. This deficiency makes it difficult to fully understand the scope and details of bacterial resistance in different regions and healthcare settings across the country. Therefore, conducting epidemiological studies in each geographic area is essential to gather accurate data, clearly demonstrate local resistance patterns, and develop strategies to effectively control the spread of resistant bacteria. The ultimate benefit of such surveillance programs lies in preventing the wider distribution of antimicrobial resistance within that specific region, thereby protecting both public health and the effectiveness of current antibiotic treatments [3, 5-7].
The currently available data predominantly focus on isolated outbreaks and particularly severe cases of resistant bacterial infections. Unfortunately, this limited scope does not provide sufficient information regarding the broader epidemiological trends, transmission dynamics, or established patterns of resistance within the population. Therefore, laboratory reports that systematically document the existing patterns of pathogens responsible for infections gain significant importance. These detailed microbiological surveillance reports play a crucial role in informing and guiding clinicians and healthcare providers. They enable healthcare teams to select the most effective and targeted treatments for managing drug-resistant infections. Additionally, such an evidence-based framework helps reduce the likelihood of further development and spread of antibiotic resistance by promoting the rational and precise use of antimicrobial agents, ultimately contributing to better patient outcomes and the preservation of public health [8-11].
Hence, this study aimed to evaluate the antibiotic susceptibility and resistance patterns among the body fluids and urine cultures of ICU patients in a tertiary care center in Tehran, Iran.
Instrument and Methods
In this cross-sectional study, we used data from body fluids (including pleural and ascitic fluid) and urine samples of ICU patients at Imam Khomeini Hospital, Tehran University of Medical Sciences, Tehran, Iran from 2019 to 2022. This hospital is affiliated with the top-ranked medical school in Iran and serves as a referral center. Consequently, numerous patients from different socio-economic backgrounds are referred to this hospital. Thus, the data can be viewed as a viable depiction of antibiotic susceptibility and resistance in the general population of Iran. Informed consent was obtained from all the patients whose data were used.
All individuals who contributed samples to this study voluntarily provided their informed consent to participate. They were thoroughly briefed on the research objectives and potential benefits. Participants were guaranteed the right to withdraw at any time without facing any repercussions. Written consent was obtained from all participants before their inclusion in the study. This study was conducted in accordance with the ethical principles outlined in the Declaration of Helsinki.
The body fluids and urine cultures were collected according to the currently available standard protocols [3, 7, 12-14]. After obtaining the samples, all types of microorganisms and susceptibility testing were recorded. These bacteria were isolated using the streak plate method on blood agar and MacConkey agar cultures. Culture plates were incubated at 37°C for 24 hours. A culture consisting of more than 105 colonies of a particular bacterium was considered a positive culture. The incubation of negative 24-hour cultures was extended for an additional 24 hours. Species were identified by biochemical tests (indole, citrate, oxidase and H2S production, lysine decarboxylase, lactose fermentation, urea hydrolysis, gas production, catalase, coagulase, mannitol fermentation, and susceptibility testing for novobiocin).
After identification, antibiotic susceptibility and resistance were tested using the diffusion method on Mueller-Hinton medium (Merck, Germany). After inoculating the bacteria on Mueller-Hinton agar and placing antibiotic discs, the plates were incubated in an incubator for 24 hours. The results were then classified according to the size of the growth inhibition zones and international standards, and reported in three groups: 1) susceptible and sensitive, 2) intermediate susceptibility or sensitivity, and 3) resistance.
We used STATA 17 to analyze and report the results.
Findings
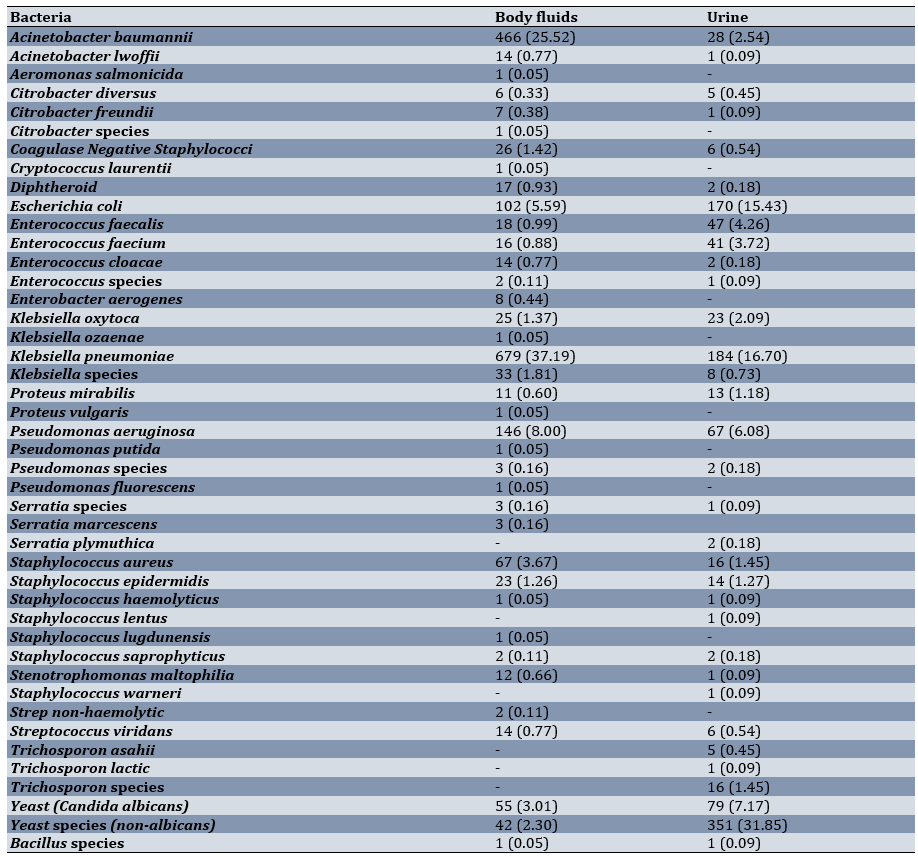
Overall, 4,232 body fluid samples and 4,062 urine samples were collected. Among the body fluid samples, 2,400 cultures were negative, and among the urine samples, 2,824 cultures were negative. Among the body fluid samples, 97 cultures (5.31%) were positive for yeast (Candida species). Among the urine samples, 430 cultures (36.94%) were positive for yeast (Candida species). The most common bacterial species found in the urine cultures were Klebsiella pneumoniae (184, 16.70%), followed by Escherichia coli (170, 15.43%) and Pseudomonas aeruginosa (67, 6.08%). The most common bacterial species found in the body fluid cultures were Klebsiella pneumoniae (679, 37.19%), followed by Acinetobacter baumannii (466, 25.52%) and Pseudomonas aeruginosa (146, 8%; Table 1).
Table 1. Frequency of different species among the body fluid and urine samples
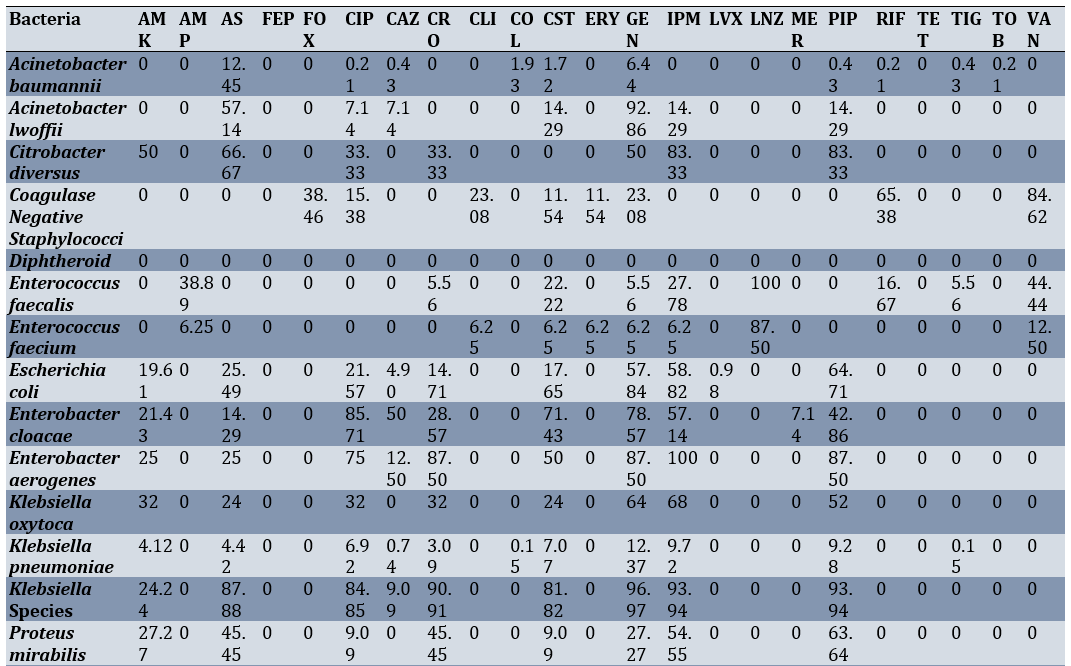
Among the body fluid samples, the highest sensitivity of Klebsiella pneumoniae was observed against gentamicin (12.37%) and imipenem (9.72%), respectively. The highest sensitivity of Escherichia coli was noted against piperacillin-tazobactam (67.71%) and imipenem (58.82%), respectively (Table 2).
Table 2. Sensitivity percentage of various bacterial species isolated from intensive care unit body fluid cultures to different antibiotics
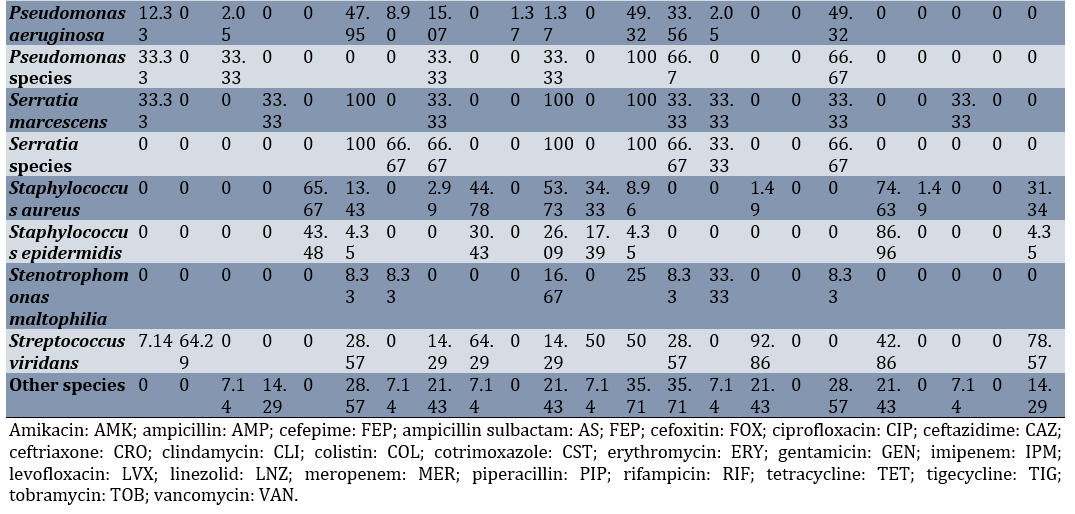
Additionally, Klebsiella pneumoniae exhibited the highest resistance to ampicillin-sulbactam (91.61%), ciprofloxacin (88.66%), and cotrimoxazole (88.37%), respectively. The highest resistance of Escherichia coli was observed against cotrimoxazole (76.47%) and ciprofloxacin (72.55%; Table 3).
Table 3. Resistance percentage of various bacterial species isolated from intensive care unit body fluid cultures to different antibiotics
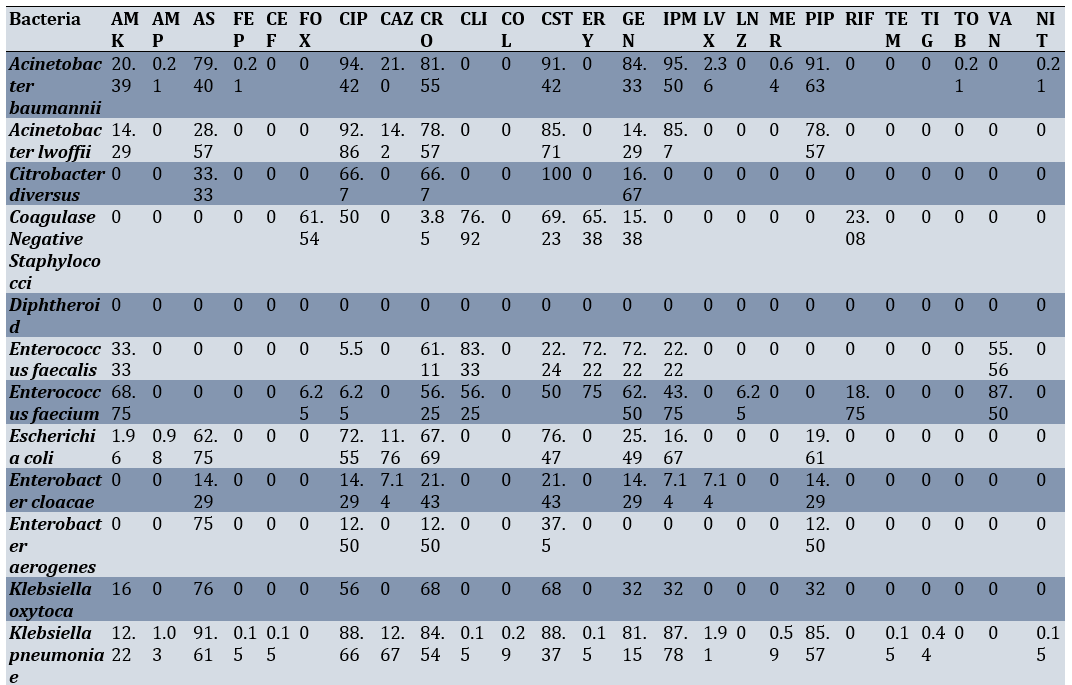
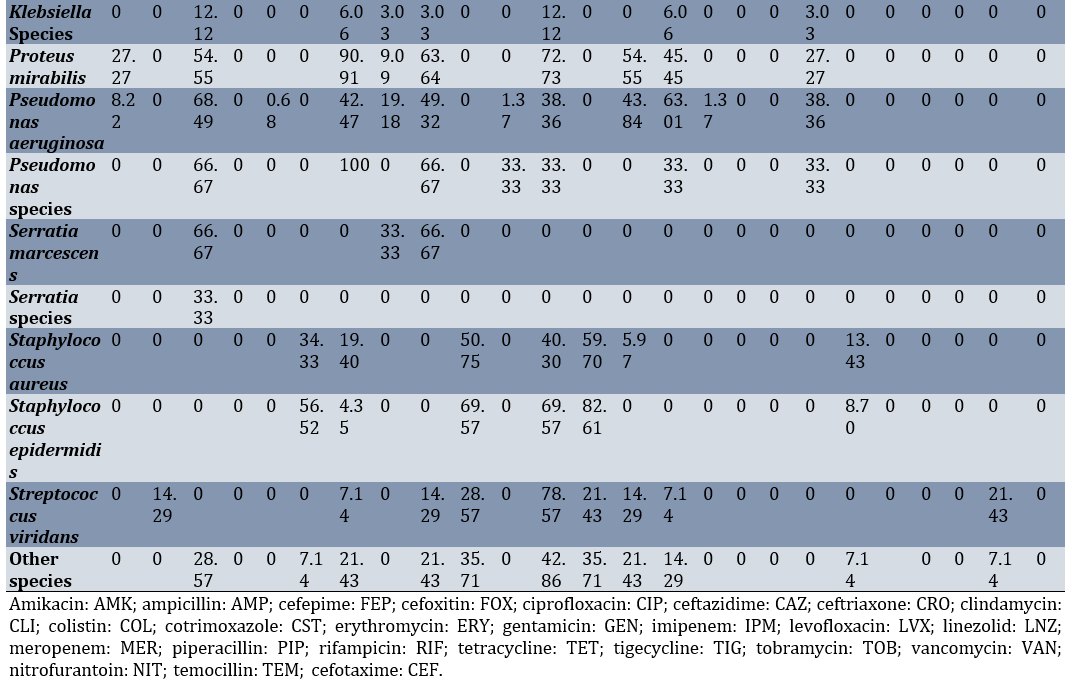
Among the urine samples, the highest sensitivity of Klebsiella pneumoniae was seen against Gentamicin (17.93%), Imipenem (14.67%), and piperacillin-tazobactam (12.50%), respectively. The highest sensitivity of Acinetobacter baumannii was recorded against imipenem (88.82%) and nitrofurantoin (88.24%; Table 4).
Table 4. Sensitivity percentage of various bacterial species isolated from intensive care unit urine cultures to different antibiotics
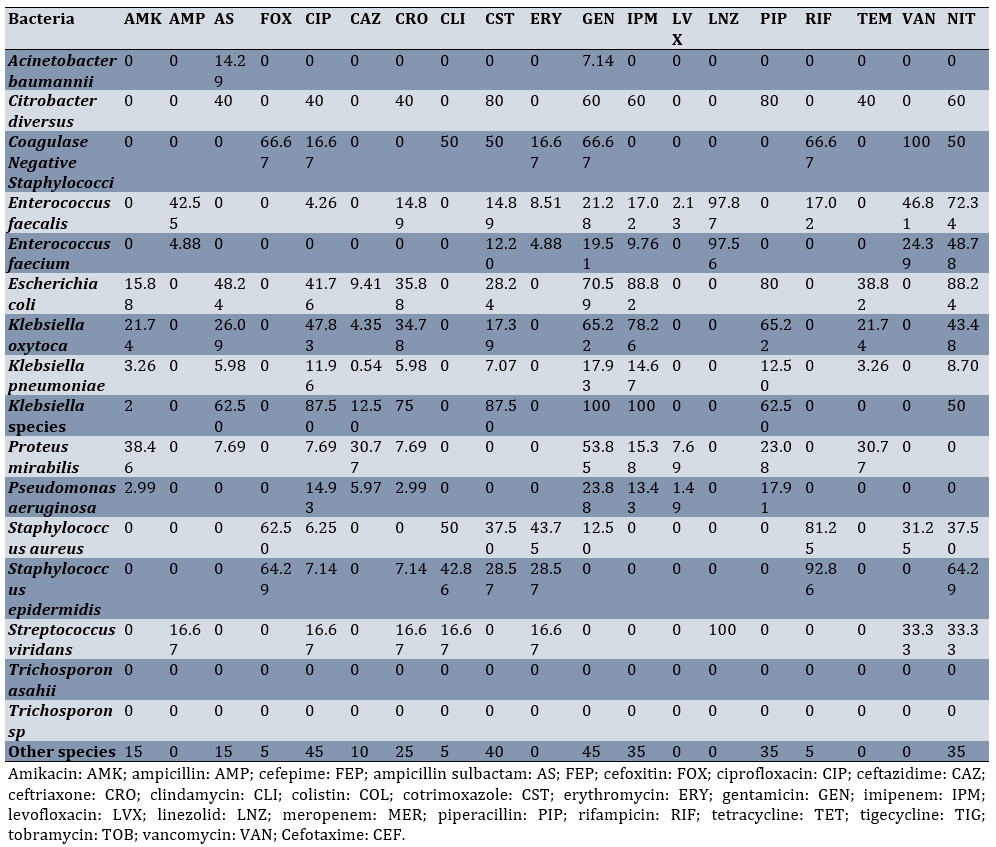
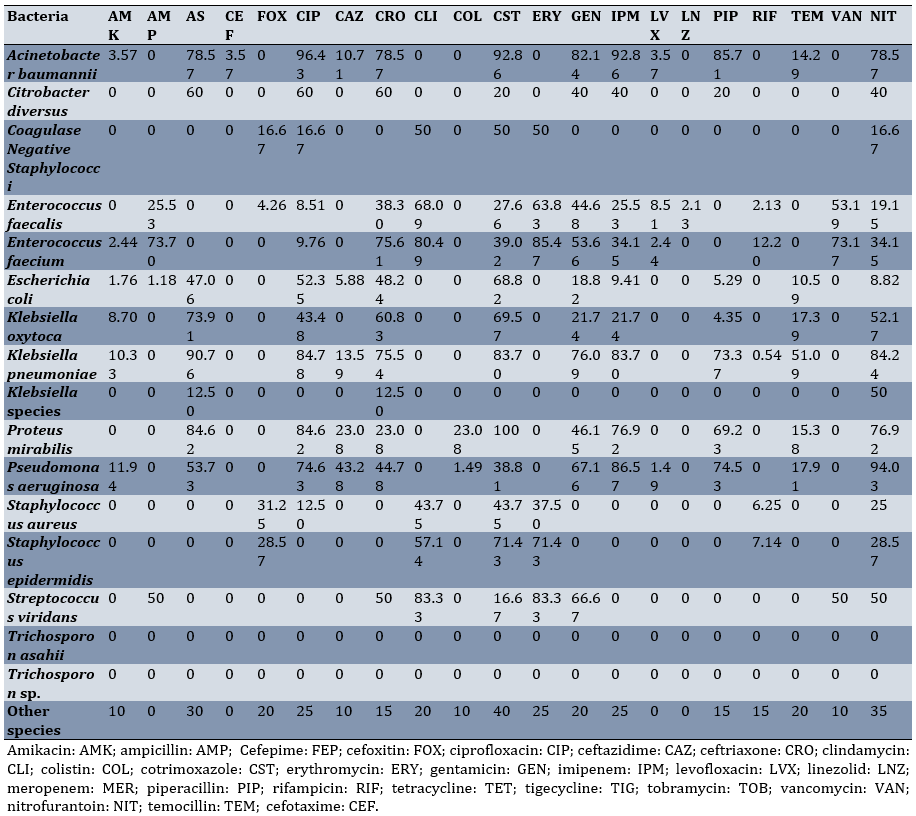
Klebsiella pneumoniae was most resistant to ampicillin-sulbactam (90.76%), ciprofloxacin (84.78%), and nitrofurantoin (84.24%), respectively. The highest resistance of Acinetobacter baumannii was noted against cotrimoxazole (68.82%) and ciprofloxacin (52.35%; Table 5; Appendix 1)
Table 5. Resistance percentage of various bacterial species isolated from intensive care unit urine cultures to different antibiotics
Discussion
The rise of bacterial co-infection or secondary infection among ICU-admitted patients has become alarming globally. Bacterial infection is one of the most severe complications faced by critically ill patients admitted to the ICU. Proper antibiotic therapy requires precise and timely isolation, identification, and performance of sensitivity tests. Inappropriate or delayed antibiotic treatment may increase the chances of antibiotic resistance, prolonged infection, and increased mortality in ICU patients. Intrahospital surveillance programs are necessary to identify these agents and provide appropriate treatment. Such studies can inform us about the changing patterns of antibiotic resistance and susceptibility [15-17].
Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii were the most prevalent bacterial species found in the collected cultures. All of these species exhibited diminished antibiotic susceptibility and increased antibiotic resistance, which should be concerning at both national and international levels. This claim is supported by numerous studies conducted over the last decade, both in Iran and internationally. According to a recent study, Klebsiella pneumoniae, Staphylococcus aureus, and Acinetobacter baumannii are the most prevalent bacterial species collected from critically ill patients with secondary infections. These findings align with the results of our study [4, 18-21].
In a study designed to assess changes in antibiotic resistance patterns in Iran, Khoshbakht et al. reported that the bacterial species with the highest antibiotic resistance are Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii [22]. These species are resistant to ampicillin, imipenem, and ceftazidime. Similar results were obtained in our study, as Acinetobacter baumannii was also resistant to imipenem. According to this study, Klebsiella pneumoniae has shown a rising resistance, reaching 30% over the course of one year, to a wide spectrum of antibiotics. Another study reports an increase in the resistance of Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae during the COVID-19 outbreak in Iran, with resistance levels reaching up to 89% for multiple antibiotics. According to the results of our study, Escherichia coli was resistant to more than 80% of imipenem and nitrofurantoin, which is consistent with other reports [23-30].
Another study indicates a rising trend in antibiotic resistance of Escherichia coli against beta-lactams, aminoglycosides, quinolones, tetracycline, chloramphenicol, and cotrimoxazole [31]. Although the pattern of resistance varies from region to region in Iran, the prevalence of Escherichia coli infection is significantly high in more than half of the provinces in the country. There is a lack of evidence regarding the prevalence and resistance patterns of Escherichia coli in other provinces of Iran [31]. A similar study conducted on patients in a hospital in Tehran reports that colistin and tigecycline are the most effective antibiotic therapy options against Escherichia coli [32-34].
Our results suggest a high resistance among prevalent bacterial species, such as Klebsiella pneumoniae, Acinetobacter baumannii, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus against a variety of antimicrobial agents. Such findings should be alarming for policymakers and necessitate the implementation of active surveillance programs in hospitals, especially in ICU wards, to monitor the prevalence, susceptibility, and resistance of these infectious microorganisms.
Conclusion
Klebsiella pneumoniae, Acinetobacter baumannii, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus have a high resistance against many antimicrobial agents.
Acknowledgments: None.
Ethical Permissions: Ethical approval was obtained from the ethics committee of Tehran University of Medical Sciences.
Conflicts of Interests: The authors declared no conflicts of interests.
Authors' Contribution: Nazar E (First Author), Introduction Writer/Main Researcher (20%); Abdollahi A (Second Author), Methodologist (10%); Afarinesh Khaki P (Third Author), Introduction Writer (10%); Norouzi Shadehi M (Fourth Author), Statistical Analyst (10%); Beighmohammadi MT (Fifth Author), Discussion Writer (10%); Nateghi S (Sixth Author), Statistical Analyst (10%); Panahi Z (Seventh Author), Discussion Writer (10%); Nozheh A (Eighth Author), Introduction Writer/Assistant Researcher (20%)
Funding/Support: We received no funding.
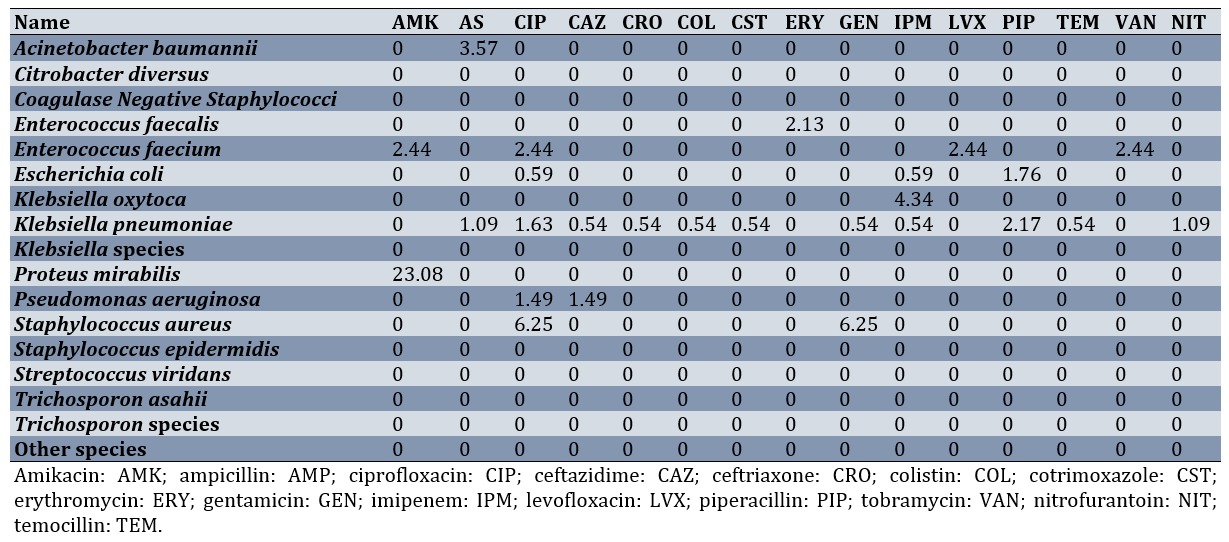
Appendix 1. ICU Urine Cultures-Intermediate sensitivity to antibiotics
Antibiotic resistance has become one of the most important determinants of outcomes in patients with severe infections. A recent estimate reports that more than ten million patients can die each year globally from antibiotic resistance. This rising number reflects a growing public health crisis that threatens to undermine the effectiveness of existing antibiotic treatments. The report emphasizes that global action is necessary to tackle this issue efficiently and sustainably. The impact of such a response is particularly relevant in settings with high rates of multidrug resistance, such as hospitals and, more importantly, intensive care units (ICUs). These critical care units are especially vulnerable due to factors like the frequent use of broad-spectrum antibiotics, prolonged patient stays, invasive procedures, and patients’ compromised immune statuses. In these environments, antibiotic-resistant bacteria can spread rapidly, complicate treatment regimens, and significantly increase mortality rates. Without effective interventions, including enhanced surveillance, antibiotic stewardship programs, and infection prevention and control measures, the burden of antibiotic resistance in ICUs will continue to rise, threatening patient outcomes and healthcare systems worldwide [1-4].
National action plans and surveillance are indeed necessary to mitigate this issue. The problem of antibiotic resistance is an emerging and serious challenge worldwide. Unfortunately, Iran still lacks a well-organized national surveillance system to comprehensively collect and monitor data on antimicrobial resistance and antibiotic susceptibility. This deficiency makes it difficult to fully understand the scope and details of bacterial resistance in different regions and healthcare settings across the country. Therefore, conducting epidemiological studies in each geographic area is essential to gather accurate data, clearly demonstrate local resistance patterns, and develop strategies to effectively control the spread of resistant bacteria. The ultimate benefit of such surveillance programs lies in preventing the wider distribution of antimicrobial resistance within that specific region, thereby protecting both public health and the effectiveness of current antibiotic treatments [3, 5-7].
The currently available data predominantly focus on isolated outbreaks and particularly severe cases of resistant bacterial infections. Unfortunately, this limited scope does not provide sufficient information regarding the broader epidemiological trends, transmission dynamics, or established patterns of resistance within the population. Therefore, laboratory reports that systematically document the existing patterns of pathogens responsible for infections gain significant importance. These detailed microbiological surveillance reports play a crucial role in informing and guiding clinicians and healthcare providers. They enable healthcare teams to select the most effective and targeted treatments for managing drug-resistant infections. Additionally, such an evidence-based framework helps reduce the likelihood of further development and spread of antibiotic resistance by promoting the rational and precise use of antimicrobial agents, ultimately contributing to better patient outcomes and the preservation of public health [8-11].
Hence, this study aimed to evaluate the antibiotic susceptibility and resistance patterns among the body fluids and urine cultures of ICU patients in a tertiary care center in Tehran, Iran.
Instrument and Methods
In this cross-sectional study, we used data from body fluids (including pleural and ascitic fluid) and urine samples of ICU patients at Imam Khomeini Hospital, Tehran University of Medical Sciences, Tehran, Iran from 2019 to 2022. This hospital is affiliated with the top-ranked medical school in Iran and serves as a referral center. Consequently, numerous patients from different socio-economic backgrounds are referred to this hospital. Thus, the data can be viewed as a viable depiction of antibiotic susceptibility and resistance in the general population of Iran. Informed consent was obtained from all the patients whose data were used.
All individuals who contributed samples to this study voluntarily provided their informed consent to participate. They were thoroughly briefed on the research objectives and potential benefits. Participants were guaranteed the right to withdraw at any time without facing any repercussions. Written consent was obtained from all participants before their inclusion in the study. This study was conducted in accordance with the ethical principles outlined in the Declaration of Helsinki.
The body fluids and urine cultures were collected according to the currently available standard protocols [3, 7, 12-14]. After obtaining the samples, all types of microorganisms and susceptibility testing were recorded. These bacteria were isolated using the streak plate method on blood agar and MacConkey agar cultures. Culture plates were incubated at 37°C for 24 hours. A culture consisting of more than 105 colonies of a particular bacterium was considered a positive culture. The incubation of negative 24-hour cultures was extended for an additional 24 hours. Species were identified by biochemical tests (indole, citrate, oxidase and H2S production, lysine decarboxylase, lactose fermentation, urea hydrolysis, gas production, catalase, coagulase, mannitol fermentation, and susceptibility testing for novobiocin).
After identification, antibiotic susceptibility and resistance were tested using the diffusion method on Mueller-Hinton medium (Merck, Germany). After inoculating the bacteria on Mueller-Hinton agar and placing antibiotic discs, the plates were incubated in an incubator for 24 hours. The results were then classified according to the size of the growth inhibition zones and international standards, and reported in three groups: 1) susceptible and sensitive, 2) intermediate susceptibility or sensitivity, and 3) resistance.
We used STATA 17 to analyze and report the results.
Findings
Overall, 4,232 body fluid samples and 4,062 urine samples were collected. Among the body fluid samples, 2,400 cultures were negative, and among the urine samples, 2,824 cultures were negative. Among the body fluid samples, 97 cultures (5.31%) were positive for yeast (Candida species). Among the urine samples, 430 cultures (36.94%) were positive for yeast (Candida species). The most common bacterial species found in the urine cultures were Klebsiella pneumoniae (184, 16.70%), followed by Escherichia coli (170, 15.43%) and Pseudomonas aeruginosa (67, 6.08%). The most common bacterial species found in the body fluid cultures were Klebsiella pneumoniae (679, 37.19%), followed by Acinetobacter baumannii (466, 25.52%) and Pseudomonas aeruginosa (146, 8%; Table 1).
Table 1. Frequency of different species among the body fluid and urine samples

Among the body fluid samples, the highest sensitivity of Klebsiella pneumoniae was observed against gentamicin (12.37%) and imipenem (9.72%), respectively. The highest sensitivity of Escherichia coli was noted against piperacillin-tazobactam (67.71%) and imipenem (58.82%), respectively (Table 2).
Table 2. Sensitivity percentage of various bacterial species isolated from intensive care unit body fluid cultures to different antibiotics


Additionally, Klebsiella pneumoniae exhibited the highest resistance to ampicillin-sulbactam (91.61%), ciprofloxacin (88.66%), and cotrimoxazole (88.37%), respectively. The highest resistance of Escherichia coli was observed against cotrimoxazole (76.47%) and ciprofloxacin (72.55%; Table 3).
Table 3. Resistance percentage of various bacterial species isolated from intensive care unit body fluid cultures to different antibiotics


Among the urine samples, the highest sensitivity of Klebsiella pneumoniae was seen against Gentamicin (17.93%), Imipenem (14.67%), and piperacillin-tazobactam (12.50%), respectively. The highest sensitivity of Acinetobacter baumannii was recorded against imipenem (88.82%) and nitrofurantoin (88.24%; Table 4).
Table 4. Sensitivity percentage of various bacterial species isolated from intensive care unit urine cultures to different antibiotics

Klebsiella pneumoniae was most resistant to ampicillin-sulbactam (90.76%), ciprofloxacin (84.78%), and nitrofurantoin (84.24%), respectively. The highest resistance of Acinetobacter baumannii was noted against cotrimoxazole (68.82%) and ciprofloxacin (52.35%; Table 5; Appendix 1)
Table 5. Resistance percentage of various bacterial species isolated from intensive care unit urine cultures to different antibiotics

Discussion
The rise of bacterial co-infection or secondary infection among ICU-admitted patients has become alarming globally. Bacterial infection is one of the most severe complications faced by critically ill patients admitted to the ICU. Proper antibiotic therapy requires precise and timely isolation, identification, and performance of sensitivity tests. Inappropriate or delayed antibiotic treatment may increase the chances of antibiotic resistance, prolonged infection, and increased mortality in ICU patients. Intrahospital surveillance programs are necessary to identify these agents and provide appropriate treatment. Such studies can inform us about the changing patterns of antibiotic resistance and susceptibility [15-17].
Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa, and Acinetobacter baumannii were the most prevalent bacterial species found in the collected cultures. All of these species exhibited diminished antibiotic susceptibility and increased antibiotic resistance, which should be concerning at both national and international levels. This claim is supported by numerous studies conducted over the last decade, both in Iran and internationally. According to a recent study, Klebsiella pneumoniae, Staphylococcus aureus, and Acinetobacter baumannii are the most prevalent bacterial species collected from critically ill patients with secondary infections. These findings align with the results of our study [4, 18-21].
In a study designed to assess changes in antibiotic resistance patterns in Iran, Khoshbakht et al. reported that the bacterial species with the highest antibiotic resistance are Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii [22]. These species are resistant to ampicillin, imipenem, and ceftazidime. Similar results were obtained in our study, as Acinetobacter baumannii was also resistant to imipenem. According to this study, Klebsiella pneumoniae has shown a rising resistance, reaching 30% over the course of one year, to a wide spectrum of antibiotics. Another study reports an increase in the resistance of Escherichia coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae during the COVID-19 outbreak in Iran, with resistance levels reaching up to 89% for multiple antibiotics. According to the results of our study, Escherichia coli was resistant to more than 80% of imipenem and nitrofurantoin, which is consistent with other reports [23-30].
Another study indicates a rising trend in antibiotic resistance of Escherichia coli against beta-lactams, aminoglycosides, quinolones, tetracycline, chloramphenicol, and cotrimoxazole [31]. Although the pattern of resistance varies from region to region in Iran, the prevalence of Escherichia coli infection is significantly high in more than half of the provinces in the country. There is a lack of evidence regarding the prevalence and resistance patterns of Escherichia coli in other provinces of Iran [31]. A similar study conducted on patients in a hospital in Tehran reports that colistin and tigecycline are the most effective antibiotic therapy options against Escherichia coli [32-34].
Our results suggest a high resistance among prevalent bacterial species, such as Klebsiella pneumoniae, Acinetobacter baumannii, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus against a variety of antimicrobial agents. Such findings should be alarming for policymakers and necessitate the implementation of active surveillance programs in hospitals, especially in ICU wards, to monitor the prevalence, susceptibility, and resistance of these infectious microorganisms.
Conclusion
Klebsiella pneumoniae, Acinetobacter baumannii, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus have a high resistance against many antimicrobial agents.
Acknowledgments: None.
Ethical Permissions: Ethical approval was obtained from the ethics committee of Tehran University of Medical Sciences.
Conflicts of Interests: The authors declared no conflicts of interests.
Authors' Contribution: Nazar E (First Author), Introduction Writer/Main Researcher (20%); Abdollahi A (Second Author), Methodologist (10%); Afarinesh Khaki P (Third Author), Introduction Writer (10%); Norouzi Shadehi M (Fourth Author), Statistical Analyst (10%); Beighmohammadi MT (Fifth Author), Discussion Writer (10%); Nateghi S (Sixth Author), Statistical Analyst (10%); Panahi Z (Seventh Author), Discussion Writer (10%); Nozheh A (Eighth Author), Introduction Writer/Assistant Researcher (20%)
Funding/Support: We received no funding.
Appendix 1. ICU Urine Cultures-Intermediate sensitivity to antibiotics

Keywords:
Antibiotic Resistance [MeSH], Urine [MeSH], Antibiogram [MeSH], Intensive Care Unit [MeSH], Iran [MeSH]
References
1. Van Duijn PJ, Verbrugghe W, Jorens PG, Spöhr F, Schedler D, Deja M, et al. The effects of antibiotic cycling and mixing on acquisition of antibiotic resistant bacteria in the ICU: A post-hoc individual patient analysis of a prospective cluster-randomized crossover study. PLoS One. 2022;17(5):e0265720. [Link] [DOI:10.1371/journal.pone.0265720]
2. Serra-Burriel M, Campillo-Artero C, Agodi A, Barchitta M, López-Casasnovas G. Association between antibiotic resistance in intensive care unit (ICU)-acquired infections and excess resource utilization: Evidence from Spain, Italy, and Portugal. Infect Control Hosp Epidemiol. 2022;43(10):1360-7. [Link] [DOI:10.1017/ice.2021.429]
3. Cornes M, Ibarz M, Ivanov H, Grankvist K. Blood sampling guidelines with focus on patient safety and identification-a review. Diagnosis. 2019;6(1):33-7. [Link] [DOI:10.1515/dx-2018-0042]
4. De Waele JJ, Akova M, Antonelli M, Canton R, Carlet J, De Backer D, et al. Antimicrobial resistance and antibiotic stewardship programs in the ICU: Insistence and persistence in the fight against resistance. A position statement from ESICM/ESCMID/WAAAR round table on multi-drug resistance. Intensive Care Med. 2018;44(2):189-96. [Link] [DOI:10.1007/s00134-017-5036-1]
5. Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; A critical review. Genes Dis. 2019;6(2):109-19. [Link] [DOI:10.1016/j.gendis.2019.04.001]
6. Weber DJ, Raasch R, Rutala WA. Nosocomial infections in the ICU: The growing importance of antibiotic-resistant pathogens. Chest. 1999;115(3):34S-41S. [Link] [DOI:10.1378/chest.115.suppl_1.34S]
7. Moradi Tabriz H, Abdollahi A, Mahfoozi S. Frequency of pathogens and antimicrobial susceptibility of bacteria isolated from bloodstream infections. Iran J Pathol. 2010;5(3):143-9. [Link]
8. Hanberger H, Diekema D, Fluit A, Jones R, Struelens M, Spencer R, et al. Surveillance of antibiotic resistance in European ICUs. J Hosp Infect. 2001;48(3):161-76. [Link] [DOI:10.1053/jhin.2001.0987]
9. Oostdijk EA, Kesecioglu J, Schultz MJ, Visser CE, De Jonge E, Van Essen EH, et al. Effects of decontamination of the oropharynx and intestinal tract on antibiotic resistance in ICUs: A randomized clinical trial. JAMA. 2014;312(14):1429-37. [Link] [DOI:10.1001/jama.2014.7247]
10. Nerurkar A, Solanky P, Naik SS. Bacterial pathogens in urinary tract infection and antibiotic susceptibility pattern. J Pharm Biomed Sci. 2012;21(21). [Link]
11. Shrestha S, Shrestha N, Singh SD, Shrestha R, Kayestha S, Shrestha M, et al. Bacterial isolates and its antibiotic susceptibility pattern in NICU. Kathmandu Univ Med J. 2013;11(41):66-70. [Link] [DOI:10.3126/kumj.v11i1.11030]
12. Ochoa Sangrador C, Brezmes Valdivieso MF; Grupo Investigador del Proyecto. Sample collection methods for urine culture and analysis. An Pediatr. 2007;67(5):442-9. [Spanish] [Link] [DOI:10.1157/13111597]
13. Thomas V, Clark J, Doré J. Fecal microbiota analysis: An overview of sample collection methods and sequencing strategies. Future Microbiol. 2015;10(9):1485-504. [Link] [DOI:10.2217/fmb.15.87]
14. Nozarian Z, Abdollahi A. Microbial etiology and antimicrobial susceptibility of bactria implicated in urinary tract infection in Tehran, Iran. Iran J Pathol. 2015;10(1):54-60. [Link]
15. Plantinga NL, Wittekamp BH, Van Duijn PJ, Bonten MJ. Fighting antibiotic resistance in the intensive care unit using antibiotics. Future Microbiol. 2015;10(3):391-406. [Link] [DOI:10.2217/fmb.14.146]
16. Barnsteiner S, Baty F, Albrich WC, Babouee Flury B, Gasser M, Plüss-Suard C, et al. Antimicrobial resistance and antibiotic consumption in intensive care units, Switzerland, 2009 to 2018. Euro Surveill. 2021;26(46):2001537. [Link] [DOI:10.2807/1560-7917.ES.2021.26.46.2001537]
17. Kollef MH, Fraser VJ. Antibiotic resistance in the intensive care unit. Ann Intern Med. 2001;134(4):298-314. [Link] [DOI:10.7326/0003-4819-134-4-200102200-00014]
18. Lee WC, Ho MC, Leu SW, Chang CC, Lin CK, Lin CM, et al. The impacts of bacterial co-infections and secondary bacterial infections on patients with severe influenza pneumonitis admitted to the intensive care units. J Crit Care. 2022;72:154164. [Link] [DOI:10.1016/j.jcrc.2022.154164]
19. Deege MP, Paterson DL. Reducing the development of antibiotic resistance in critical care units. Curr Pharm Biotechnol. 2011;12(12):2062-9. [Link] [DOI:10.2174/138920111798808301]
20. Lepape A, Jean A, De Waele J, Friggeri A, Savey A, Vanhems P, et al. European intensive care physicians' experience of infections due to antibiotic-resistant bacteria. Antimicrob Resist Infect Control. 2020;9(1):1. [Link] [DOI:10.1186/s13756-019-0662-8]
21. Saharman YR, Karuniawati A, Severin JA, Verbrugh HA. Infections and antimicrobial resistance in intensive care units in lower-middle income countries: A scoping review. Antimicrob Resist Infect Control. 2021;10(1):22. [Link] [DOI:10.1186/s13756-020-00871-x]
22. Khoshbakht R, Kabiri M, Neshani A, Khaksari MN, Sadrzadeh SM, Mousavi SM, et al. Assessment of antibiotic resistance changes during the Covid-19 pandemic in northeast of Iran during 2020-2022: An epidemiological study. Antimicrob Resist Infect Control. 2022;11(1):121. [Link] [DOI:10.1186/s13756-022-01159-y]
23. Raoofi R, Namavari N, Rahmanian V, Dousthaghi MH. Evaluation of antibiotics resistance in Southern Iran in light of COVID-19 pandemic: A retrospective observational study. Health Sci Rep. 2023;6(3):e1153. [Link] [DOI:10.1002/hsr2.1153]
24. Mehtarpour M, Takian A, Eshrati B, Jaafaripooyan E. Control of antimicrobial resistance in Iran: The role of international factors. BMC Public Health. 2020;20(1):873. [Link] [DOI:10.1186/s12889-020-09006-8]
25. Moradi J, Hashemi FB, Bahador A. Antibiotic resistance of Acinetobacter baumannii in Iran: A systemic review of the published literature. Osong Public Health Res Perspect. 2015;6(2):79-86. [Link] [DOI:10.1016/j.phrp.2014.12.006]
26. Heidary M, Nasiri MJ, Dabiri H, Tarashi S. Prevalence of drug-resistant klebsiella pneumoniae in Iran: A review article. Iran J Public Health. 2018;47(3):317-26. [Link]
27. Shoaee S, Rezaie F, Payab M, Bakhtiari F, Heydari MH. Experiences from the management of COVID-19 pandemic in a nursing home in Iran (March-April, 2020). J Diabetes Metab Disord. 2022;21(1):1195-9. [Link] [DOI:10.1007/s40200-022-01005-3]
28. Sofi-Mahmudi A, Masinaei M, Shamsoddin E, Tovani-Palone MR, Heydari MH, Shoaee S, et al. Global, regional, and national burden and quality of care index (QCI) of lip and oral cavity cancer: A systematic analysis of the global burden of disease study 1990-2017. BMC Oral Health. 2021;21(1):558. [Link] [DOI:10.1186/s12903-021-01918-0]
29. Shoaee S, Saeedi Moghaddam S, Masinaei M, Sofi-Mahmudi A, Hessari H, Heydari MH, et al. Trends in dental caries of deciduous teeth in Iran: A systematic analysis of the national and sub-national data from 1990 to 2017. BMC Oral Health. 2022;22(1):634. [Link] [DOI:10.1186/s12903-022-02634-z]
30. Mortazavi H, Sadeghian A, Hazrati P, Heydari MH, Madihi S. Oral hemorrhagic blister and its possible related factors: Analyzes of reported cases in the literature. J Oral Maxillofac Surg Med Pathol. 2023;35(4):358-67. [Link] [DOI:10.1016/j.ajoms.2022.12.009]
31. Alizade H. Escherichia coli in Iran: An overview of antibiotic resistance: A review article. Iran J Public Health. 2018;47(1):1-12. [Link]
32. Sharahi JY, Hashemi A, Ardebili A, Davoudabadi S. Molecular characteristics of antibiotic-resistant Escherichia coli and Klebsiella pneumoniae strains isolated from hospitalized patients in Tehran, Iran. Ann Clin Microbiol Antimicrob. 2021;20(1):32. [Link] [DOI:10.1186/s12941-021-00437-8]
33. Karimi E, Ghalibafan F, Esfandani A, Manoochehri Arash N, Mohammadi S, Khaledi A, et al. Antibiotic resistance pattern in pseudomonas aeruginosa isolated from clinical samples other than burn samples in Iran. Avicenna J Med Biotechnol. 2021;13(1):35-41. [Link] [DOI:10.18502/ajmb.v13i1.4575]
34. Vaez H, Salehi-Abargouei A, Ghalehnoo ZR, Khademi F. Multidrug resistant pseudomonas aeruginosa in Iran: A systematic review and metaanalysis. J Glob Infect Dis. 2018;10(4):212-7. [Link] [DOI:10.4103/jgid.jgid_113_17]








