Volume 5, Issue 3 (2024)
J Clinic Care Skill 2024, 5(3): 151-156 |
Back to browse issues page
Article Type:
Subject:
Ethics code: IR.YUMS.REC.1399.096
History
Received: 2024/07/12 | Accepted: 2024/09/3 | Published: 2024/09/23
Received: 2024/07/12 | Accepted: 2024/09/3 | Published: 2024/09/23
How to cite this article
Rrashidpour F, Mazloomirad F, Khoramrooz S, Sharifi A, Ghatee M, Khosravani S, et al . Antibiotic Susceptibility and Frequency of Plasmid Mediated Quinolones Resistant (qnr) Genes amongst Urine Klebsiella Pneumoniae Isolates. J Clinic Care Skill 2024; 5 (3) :151-156
URL: http://jccs.yums.ac.ir/article-1-284-en.html
URL: http://jccs.yums.ac.ir/article-1-284-en.html
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



Rights and permissions
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Authors
F. Rrashidpour1 
 , F. Mazloomirad1
, F. Mazloomirad1 
 , S.S. Khoramrooz2
, S.S. Khoramrooz2 
 , A. Sharifi2
, A. Sharifi2 
 , M.A. Ghatee2
, M.A. Ghatee2 
 , S.R. Khosravani3
, S.R. Khosravani3 
 , A. Mansourian4
, A. Mansourian4 
 , S.A. Khosravani *2
, S.A. Khosravani *2 


 , F. Mazloomirad1
, F. Mazloomirad1 
 , S.S. Khoramrooz2
, S.S. Khoramrooz2 
 , A. Sharifi2
, A. Sharifi2 
 , M.A. Ghatee2
, M.A. Ghatee2 
 , S.R. Khosravani3
, S.R. Khosravani3 
 , A. Mansourian4
, A. Mansourian4 
 , S.A. Khosravani *2
, S.A. Khosravani *2 

1- Student Research Committee, Yasuj University of Medical Sciences, Yasuj, Iran
2- Cellular and Molecular Research Center, Yasuj University of Medical Sciences, Yasuj, Iran
3- Faculty of Dentistry, Shahid Beheshti University of Medical Sciences, Tehran, Iran
4- Department of Anesthesia, Yasuj University of Medical Sciences, Yasuj, Iran
2- Cellular and Molecular Research Center, Yasuj University of Medical Sciences, Yasuj, Iran
3- Faculty of Dentistry, Shahid Beheshti University of Medical Sciences, Tehran, Iran
4- Department of Anesthesia, Yasuj University of Medical Sciences, Yasuj, Iran
Full-Text (HTML) (841 Views)
Introduction
Urinary tract infections (UTIs) encompass a range of infections affecting various parts of the urinary tract, including the bladder and kidneys [1]. These infections are among the most common nosocomial infections [2]. These infections are among the most common nosocomial infections. Bacterial pathogens are the primary cause of UTIs, with K. pneumoniae being one significant contributor. The rise of antibiotic resistance among bacteria causing UTIs is a growing concern in both developed and developing countries. Quinolones are often the first-line treatment for gram-negative infections of the urinary tract [3].
Mechanisms of resistance to quinolone drugs are as follows: A) Mutations in chromosomal genes: One mechanism of resistance arises from mutations in genes encoding one or both of the drug's target enzymes, DNA gyrase and topoisomerase IV, which play roles in bacterial replication and transcriptio. These mutations occur in the ParC, gyrB, gyrA and ParE domains, respectively, which ultimately lead to a decrease in the binding of the drug to the DNA group of the enzyme [3, 4]. B) Acquisition of plasmids carrying resistance genes: There are three main mechanisms of plasmid-mediated resistance to quinolones, described as follow:
The first mechanism is qnr proteins: These include qnrA, qnrB, qnrC, qnrD, qnrS, and qnrVC, which belong to the pentapeptide repeat family and protect DNA by inhibiting the binding of quinolones to DNA gyrase and topoisomerase IV.
The second mechanism involves the presence of aac [5]. Ib-cr protein, which produces an aminoglycosidase acetyltransferase enzyme. This enzyme causes acetylation, reducing the activity of norfloxacin and ciprofloxacin.
The third recently described mechanism involves a quinolone efflux pump mediated by the qepA protein, which reduces intracellular quinolone levels.
The qnr proteins cause resistance to quinolones by two different mechanisms: In the first case, they reduce the binding of DNA gyrase and topoisomerase 4 to DNA. In the second case, these proteins bind to DNA gyrase and topoisomerase 4 and prevent quinolones from entering the parts broken by the enzyme [6, 7]. The most important mechanism of resistance of Enterobacteriaceae species to fluoroquinolones is mutations in the bacterial chromosome [8]. It has been shown that plasmids also produce resistance to quinolones, which is increasing [9]. Plasmid-induced quinolone resistance factors cause low-level resistance to fluoroquinolones. Therefore, the identification of these factors is necessary to control resistance to these antibiotics as well as epidemiological studies. Due to the fact that these factors cannot be identified by phenotypic methods, so molecular methods such as PCR are used to identify [10]. The objectives of this study were to explore the pattern of antibiotic susceptibility and frequency of qnrA, qnrB and qnrS genes in K. pneumoniae isolates secluded from patients with infections of the urinary tract in Imam Sajjad, Yasuj and Kosar hospitals in Shiraz, in southwest Iran.
Instruments and Methods
This cross-sectional study, includes the collection of 142 isolates of K. pneumoniae from patients with infections of the urinary tract, who referred to Imam Sajjad Hospital in Yasuj and Kowsar Hospital in Shiraz in southwest Iran, 2020. After initial isolation, the bacteria were confirmed by various biochemical tests such as fermentation of glucose and lactose, motility, urea, indole, TSI, SIM and oxidase.
Sample collection
Based on the results of Kim et al.'s [7] study, p=0.405 was considered and d=0.081, and based on equation 1, the sample size was calculated as 141. These samples were collected using simple random sampling.
Equation 1:
N= (Z 1- α/2)2pq/d2
The inclusion criteria are as follows: urine samples that tested positive for the growth of Klebsiella pneumoniae. Exclusion criteria include samples that cultured bacteria other than Klebsiella pneumoniae, as well as patients with duplicate sample submissions.
Antibiotic susceptibility test
Susceptibility testing against various antibiotics was carried out through disc diffusion with regard to CLSI guidelines (2017 edition) using commercially accessible discs including Nalidixic acid (NA, 30 μg Mast, England), trimethoprim/sulfamethoxazole (SXT, 23.75 μg Mast, England), ciprofloxacin (CIP, 30 μg Mast, England), ofloxacin (OFX, 5 μg Mast, England), norfloxacin (NOR, 10 μg Mast, England), imipenem (IPM, 10 μg Mast, England). The susceptible control was K. pneumoniae ATCC 10031.
Multiplex PCR
Polymerase chain reaction: DNA extraction was performed by boiling method. Briefly, several loops of bacteria (24 h) were boiled in a microtube containing sterile distilled water for 10 min at 100°C and then centrifuged. The supernatant was kept as template DNA for PCR. Each PCR reaction mixture (25 μl) contained 5 μl DNA template, 12.5 μl Mastermix (Pishgam, Iran), and 10 pmol of each primer and 1.5 μl distilled water. PCR amplifications were carried out in a thermal cycler. The supernatant which was considered as DNA template for qnr genes (qnrA, qnrB and qnrS) was detected via PCR using specific protocols and primers (Table 1) [7]. After amplification, 10 µL of the PCR products were electrophoresed (Major Science MP300, Taiwan) on 1.5% agarose gel (Pishgam, Iran) at 90 V for 45 minutes. The PCR products were stained with Gel Stain (Pishgam, Iran). They were then visualized by Gel Documentation (Major Science, Taiwan). Using SPSS software version 20, data from the laboratory and clinical results of patients were analyzed.
Table 1. Primers used in this study

Findings
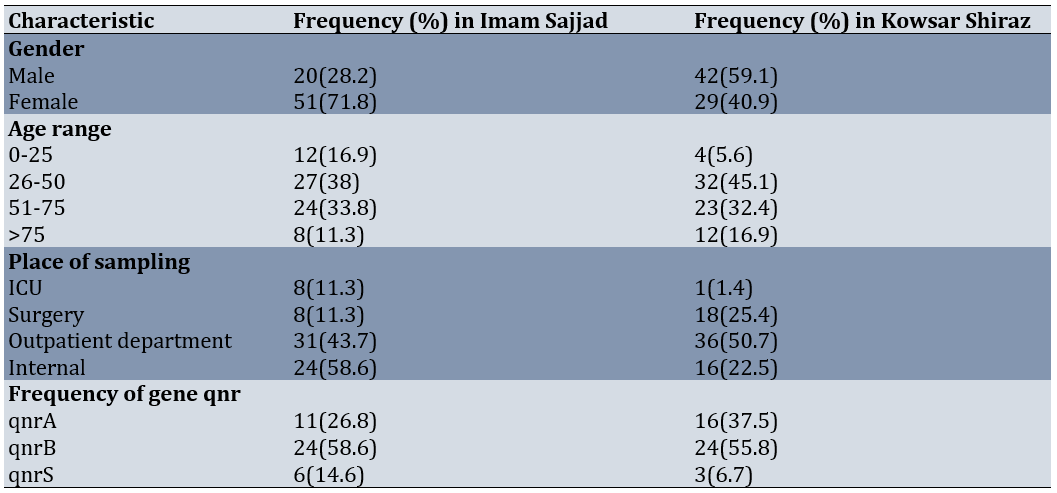
The average age of patients was 49.73±1.453 years and their age ranges were between 7 and 83 years. 80 (56%) participants were female and 62 (44%) were male. There were 71 samples (50%) from Imam Sajjad Hospital in Yasuj and 71 samples (50%) from Kosar Hospital in Shiraz. Samples taken from outpatient wards were 67 samples (47.2%), internal samples were 40 samples (28.2%), surgery samples were 26 samples (18.3%) and ICU samples were 9 samples (6.3%; Table 2).
Table 2. Characteristics of the samples taken
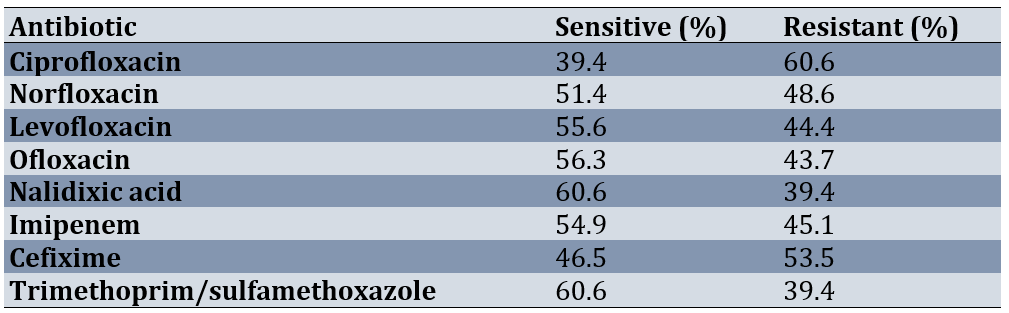
In general, K. pneumoniae isolates (142 cases) isolated from both hospitals (Imam Sajjad and Kowsar) had the highest resistance to the antibiotic ciprofloxacin 60.6%. Resistance to neurofloxacin, levofloxacin, ofloxacin and nalidixic acid was 48.6%, 44.4%, 43.7% and 39.4%, respectively. The highest susceptibility was to the antibiotics nalidixic acid and sulfamethoxazole trimethoprim. A total of 86 isolates were completely resistant to one of the types of quinolone antibiotics used in this study, which were selected to examine the presence of qnr gene assemblies using PCR. The pattern of antibiotic susceptibility of K. pneumoniae isolates was detected (Table 3).
Table 3. The pattern of antibiotic susceptibility
Results of qnr gene isolation
Out of 86 isolates insensitive to quinolones used in this study, 66 (76.7%) isolates with genes qnr plasmid-mediated isolates were identified, out of 66 isolates, 48 (55.8%) isolates containing the gene qnrB, 27 (31.4%) isolates contained qnrA gene, and 9 (10.5%) isolates carried qnrS gene (Figure 1; Figure 2).

Figure 1. Distribution of qnr genes
Urinary tract infections (UTIs) encompass a range of infections affecting various parts of the urinary tract, including the bladder and kidneys [1]. These infections are among the most common nosocomial infections [2]. These infections are among the most common nosocomial infections. Bacterial pathogens are the primary cause of UTIs, with K. pneumoniae being one significant contributor. The rise of antibiotic resistance among bacteria causing UTIs is a growing concern in both developed and developing countries. Quinolones are often the first-line treatment for gram-negative infections of the urinary tract [3].
Mechanisms of resistance to quinolone drugs are as follows: A) Mutations in chromosomal genes: One mechanism of resistance arises from mutations in genes encoding one or both of the drug's target enzymes, DNA gyrase and topoisomerase IV, which play roles in bacterial replication and transcriptio. These mutations occur in the ParC, gyrB, gyrA and ParE domains, respectively, which ultimately lead to a decrease in the binding of the drug to the DNA group of the enzyme [3, 4]. B) Acquisition of plasmids carrying resistance genes: There are three main mechanisms of plasmid-mediated resistance to quinolones, described as follow:
The first mechanism is qnr proteins: These include qnrA, qnrB, qnrC, qnrD, qnrS, and qnrVC, which belong to the pentapeptide repeat family and protect DNA by inhibiting the binding of quinolones to DNA gyrase and topoisomerase IV.
The second mechanism involves the presence of aac [5]. Ib-cr protein, which produces an aminoglycosidase acetyltransferase enzyme. This enzyme causes acetylation, reducing the activity of norfloxacin and ciprofloxacin.
The third recently described mechanism involves a quinolone efflux pump mediated by the qepA protein, which reduces intracellular quinolone levels.
The qnr proteins cause resistance to quinolones by two different mechanisms: In the first case, they reduce the binding of DNA gyrase and topoisomerase 4 to DNA. In the second case, these proteins bind to DNA gyrase and topoisomerase 4 and prevent quinolones from entering the parts broken by the enzyme [6, 7]. The most important mechanism of resistance of Enterobacteriaceae species to fluoroquinolones is mutations in the bacterial chromosome [8]. It has been shown that plasmids also produce resistance to quinolones, which is increasing [9]. Plasmid-induced quinolone resistance factors cause low-level resistance to fluoroquinolones. Therefore, the identification of these factors is necessary to control resistance to these antibiotics as well as epidemiological studies. Due to the fact that these factors cannot be identified by phenotypic methods, so molecular methods such as PCR are used to identify [10]. The objectives of this study were to explore the pattern of antibiotic susceptibility and frequency of qnrA, qnrB and qnrS genes in K. pneumoniae isolates secluded from patients with infections of the urinary tract in Imam Sajjad, Yasuj and Kosar hospitals in Shiraz, in southwest Iran.
Instruments and Methods
This cross-sectional study, includes the collection of 142 isolates of K. pneumoniae from patients with infections of the urinary tract, who referred to Imam Sajjad Hospital in Yasuj and Kowsar Hospital in Shiraz in southwest Iran, 2020. After initial isolation, the bacteria were confirmed by various biochemical tests such as fermentation of glucose and lactose, motility, urea, indole, TSI, SIM and oxidase.
Sample collection
Based on the results of Kim et al.'s [7] study, p=0.405 was considered and d=0.081, and based on equation 1, the sample size was calculated as 141. These samples were collected using simple random sampling.
Equation 1:
N= (Z 1- α/2)2pq/d2
The inclusion criteria are as follows: urine samples that tested positive for the growth of Klebsiella pneumoniae. Exclusion criteria include samples that cultured bacteria other than Klebsiella pneumoniae, as well as patients with duplicate sample submissions.
Antibiotic susceptibility test
Susceptibility testing against various antibiotics was carried out through disc diffusion with regard to CLSI guidelines (2017 edition) using commercially accessible discs including Nalidixic acid (NA, 30 μg Mast, England), trimethoprim/sulfamethoxazole (SXT, 23.75 μg Mast, England), ciprofloxacin (CIP, 30 μg Mast, England), ofloxacin (OFX, 5 μg Mast, England), norfloxacin (NOR, 10 μg Mast, England), imipenem (IPM, 10 μg Mast, England). The susceptible control was K. pneumoniae ATCC 10031.
Multiplex PCR
Polymerase chain reaction: DNA extraction was performed by boiling method. Briefly, several loops of bacteria (24 h) were boiled in a microtube containing sterile distilled water for 10 min at 100°C and then centrifuged. The supernatant was kept as template DNA for PCR. Each PCR reaction mixture (25 μl) contained 5 μl DNA template, 12.5 μl Mastermix (Pishgam, Iran), and 10 pmol of each primer and 1.5 μl distilled water. PCR amplifications were carried out in a thermal cycler. The supernatant which was considered as DNA template for qnr genes (qnrA, qnrB and qnrS) was detected via PCR using specific protocols and primers (Table 1) [7]. After amplification, 10 µL of the PCR products were electrophoresed (Major Science MP300, Taiwan) on 1.5% agarose gel (Pishgam, Iran) at 90 V for 45 minutes. The PCR products were stained with Gel Stain (Pishgam, Iran). They were then visualized by Gel Documentation (Major Science, Taiwan). Using SPSS software version 20, data from the laboratory and clinical results of patients were analyzed.
Table 1. Primers used in this study

Findings
The average age of patients was 49.73±1.453 years and their age ranges were between 7 and 83 years. 80 (56%) participants were female and 62 (44%) were male. There were 71 samples (50%) from Imam Sajjad Hospital in Yasuj and 71 samples (50%) from Kosar Hospital in Shiraz. Samples taken from outpatient wards were 67 samples (47.2%), internal samples were 40 samples (28.2%), surgery samples were 26 samples (18.3%) and ICU samples were 9 samples (6.3%; Table 2).
Table 2. Characteristics of the samples taken

In general, K. pneumoniae isolates (142 cases) isolated from both hospitals (Imam Sajjad and Kowsar) had the highest resistance to the antibiotic ciprofloxacin 60.6%. Resistance to neurofloxacin, levofloxacin, ofloxacin and nalidixic acid was 48.6%, 44.4%, 43.7% and 39.4%, respectively. The highest susceptibility was to the antibiotics nalidixic acid and sulfamethoxazole trimethoprim. A total of 86 isolates were completely resistant to one of the types of quinolone antibiotics used in this study, which were selected to examine the presence of qnr gene assemblies using PCR. The pattern of antibiotic susceptibility of K. pneumoniae isolates was detected (Table 3).
Table 3. The pattern of antibiotic susceptibility

Results of qnr gene isolation
Out of 86 isolates insensitive to quinolones used in this study, 66 (76.7%) isolates with genes qnr plasmid-mediated isolates were identified, out of 66 isolates, 48 (55.8%) isolates containing the gene qnrB, 27 (31.4%) isolates contained qnrA gene, and 9 (10.5%) isolates carried qnrS gene (Figure 1; Figure 2).

Figure 1. Distribution of qnr genes
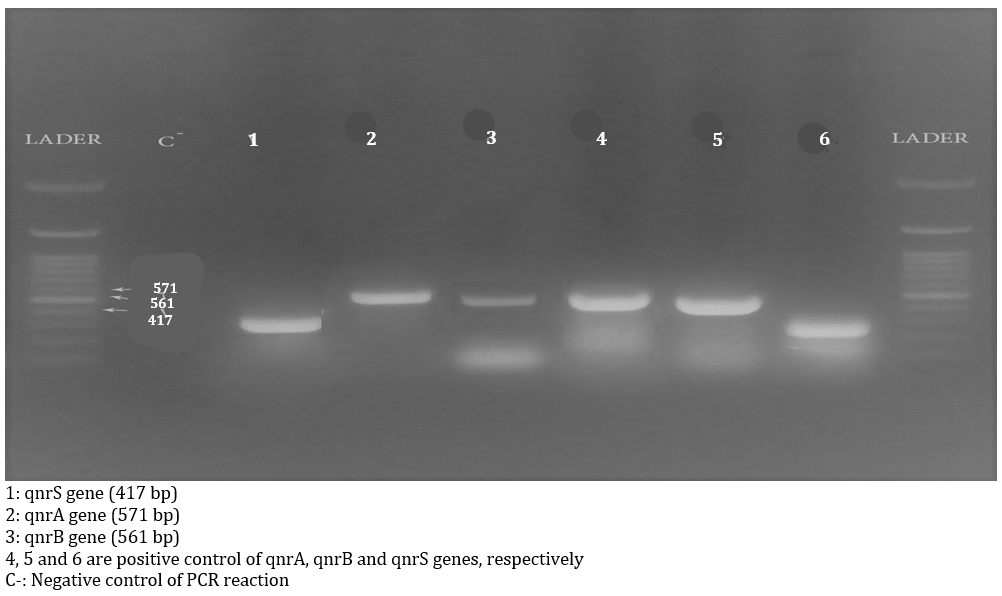
Figure 2. Gel electrophoresis image of Multiplex PCR products
Discussion
The aim of this study was to explore the pattern of antibiotic susceptibility and frequency of qnrA, qnrB and qnrS genes in K. pneumoniae isolates secluded from patients with infections of the urinary tract in Imam Sajjad, Yasuj and Kosar hospitals in Shiraz, in southwest Iran. Urinary tract infections (UTIs) comprise infections that impact multiple sections of the urinary tract, such as the bladder and kidneys. Among the primary causes of nosocomial UTIs, K. pneumoniae ranks within the top five infectious bacteria, responsible for approximately 6–17% of all UTI cases [11, 12]. The improper use of antibiotics has contributed to the rise of this pathogen as one of the most prevalent multidrug-resistant (MDR) bacteria in recent decades. Consequently, the rapid identification of MDR strains and the development of effective treatment strategies are critical. The spread of resistance factors in gram-negative bacteria has increased antimicrobial resistance, leading to prolonged infections and delayed patient recovery [12]. Quinolones are typically the first-line therapy for gram-negative UTIs [13].
In this study, resistance to nalidixic acid was observed in 39.7% of cases. This finding was lower than that reported by Azargun et al. in 2019 (100%) and by Salah et al. in 2016 (88%) [14, 15]. Despite the recent use of third-generation fluoroquinolones in Iran, 44.4% of our isolates were resistant to levofloxacin, a figure higher than the 22.6% resistance noted in the study by Nourozi et al. but lower than the 73% resistance reported by Azargun et al. [14, 18]. Ciprofloxacin showed the highest resistance level among quinolones (60.6%), aligning with findings from Ibrahim's study in Egypt and from Unlu & Demirci [16, 17]. However, this was lower compared to studies by Teimour Pour et al., Azargun et al., and Salah et al., and higher compared to results from Nourozi et al. and Mohammadbeigi et al. [13, 14, 15, 18, 19].
In 2014, Ghadami et al.'s study in Boroujerd reveal that 38% of K. pneumoniae isolates are resistant to nalidixic acid and ciprofloxacin, with resistance to ofloxacin and norfloxacin at 18% and 15%, respectively, which was lower than in the study by Nourozi et al. [18, 20]. In contrast, Teimour Pour et al. report 100% resistance to norfloxacin [13]. In this study, imipenem and sulfamethoxazole-trimethoprim showed resistance rates of 45% and 39%, respectively. These rates were lower than those found by Unlu & Demirci and Teimour Pour et al., though resistance to imipenem was lower than reported by Salah et al. [13, 15, 17].
Antibiogram findings from this study, alongside previous research, suggest that resistance rates to various antibiotics differ across studies. Contributing factors include the geographic origin of samples, patient health status and immunity, regional antibiotic use practices, healthcare policies, length of hospital stay, and overall health standards.
Although quinolone resistance has traditionally been chromosomal, plasmid-mediated resistance has recently emerged among Enterobacteriaceae, with variants such as qnr genes (qnrB, qnrA, and qnrS) becoming more prevalent worldwide [21, 22]. This study revealed a high prevalence of plasmid-mediated quinolone resistance (76.7%) among K. pneumoniae isolates resistant to quinolones. Liu et al. in China observed plasmid-mediated genes in 51% of K. pneumoniae isolates, whereas Azadpour et al. found them in 23.5% of isolates [23, 24]. Of the 86 quinolone-resistant isolates analyzed in this study, 66 (76.7%) contained qnr plasmid genes: 48 isolates (55.8%) harbored the qnrB gene, 27 (31.4%) contained the qnrA gene, and 9 (10.5%) carried the qnrS gene. In a study by Jamshidi et al. in Tehran, 25% of isolates exhibited qnrB, 18.75% show qnrS, but no isolates had qnrA [25]. Roshani et al. report resistance rates of 22.5%, 26.7%, and 46.2%, respectively [26].
A limitation of this study is that, because plasmid-mediated quinolone resistance typically results in low levels of resistance, the observed high levels of quinolone resistance in this study may suggest the involvement of additional mechanisms, such as chromosomal mutations, which were not investigated here. Although quinolone resistance is primarily due to chromosomal origins, plasmid-dependent quinolone resistance has also been increasingly reported in recent years. The findings of this study could assist in making informed preventive decisions to curb the spread of resistant strains and guide the selection of appropriate treatment protocols.
Conclusion
The highest resistance level among quinolones to be to ciprofloxacin (60.6%), while the highest antibiotic sensitivities were to sulfamethoxazole-trimethoprim (39.4%) and nalidixic acid (39.7%). Furthermore, a rising trend in antibiotic resistance among UTI pathogens is detected.
Acknowledgments: I would like to express my sincere gratitude to the cellular and molecular research center of Yasuj University of Medical Sciences for their valuable contribution and support in the preparation of this article. Their insights and feedback have greatly enriched the content of this work. I am also thankful to the microbiology department for their assistance in data analysis and research. This article would not have been possible without their help.
Ethical Permissions: This study is approved under the ethical approval code of IR.YUMS.REC.1399.096.
Conflicts of Interests: The authors declare that there is no conflict of interest.
Authors' Contribution: Rashidpour F (First Author), Original Researcher/Methodologist/Introduction Writer/Discussion Writer (20%); Mazloomirad F (Second Author), Assistant Researcher/Methodologist/Discussion Writer (10%); Khoramrooz SS (Third Author), Methodologist/Introduction Writer (10%); Sharifi A (Fourth Author), Assistant Researcher (10%); Ghatee MA (Fifth Author), Assistant Researcher (10%); Khosravani SR (Sixth Author), Assistant Researcher (10%); Mansourian A (Seventh Author), Assistant Researcher/Statistical Analyst/Discussion Writer (10%); Khosravani SA (Eighth Author), Original Researcher/Introduction Writer/Discussion Writer (20%)
Funding/Support: This work was financially supported by Yasuj University of Medical Sciences, Iran.
Keywords:
Klebsiella Pneumoniae [MeSH], Antibiotic [MeSH], Drug Resistance [MeSH], Quinolones [MeSH], Genes [MeSH]
References
1. Wright KJ, Seed PC, Hultgren SJ. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun. 2005;73(11):7657-68. [Link] [DOI:10.1128/IAI.73.11.7657-7668.2005]
2. Nielubowicz GR, Mobley HL. Host-pathogen interactions in urinary tract infection. Nat Rev Urol. 2010;7(8):430-41. [Link] [DOI:10.1038/nrurol.2010.101]
3. Jacoby GA, Strahilevitz J, Hooper DC. Plasmid‐mediated quinolone resistance. Microbiol Spectr. 2014;2(5). [Link] [DOI:10.1128/microbiolspec.PLAS-0006-2013]
4. Rajaei S, Kazemi-Pour N, Rokhbakhsh-Zamin F. Frequency of plasmid-mediated quinolone resistance genes among clinical isolates of Pseudomonas aeruginosa in Kerman, Iran. Iran J Med Microbiol. 2017;11(3):10-8. [Persian] [Link]
5. Chopra S, Galande A. A fluoroquinolone-resistant Acinetobacter baumannii without the quinolone resistance-determining region mutations. J Antimicrob Chemother. 2011;66(11):2668-70. [Link] [DOI:10.1093/jac/dkr364]
6. Cattoir V, Nordmann P. Plasmid-mediated quinolone resistance in gram-negative bacterial species: An update. Curr Med Chem. 2009;16(8):1028-46. [Link] [DOI:10.2174/092986709787581879]
7. Kim HB, Park CH, Kim CJ, Kim EC, Jacoby GA, Hooper DC. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob Agents Chemother. 2009;53(2):639-45. [Link] [DOI:10.1128/AAC.01051-08]
8. Hooper DC. Mechanisms of fluoroquinolone resistance. Drug Resist Updat. 1999;2(1):38-55. [Link] [DOI:10.1054/drup.1998.0068]
9. Stephenson S, Brown PD, Holness A, Wilks M. The emergence of qnr-mediated quinolone resistance among Enterobacteriaceae in Jamaica. West Indian Med J. 2010;59(3):241-4. [Link]
10. Pasom W, Chanawong A, Lulitanond A, Wilailuckana C, Kenprom S, Puang-Ngern P. Plasmid-mediated quinolone resistance genes, aac(6')-Ib-cr, qnrS, qnrB, and qnrA, in urinary isolates of Escherichia coli and Klebsiella pneumoniae at a teaching hospital, Thailand. Jpn J Infect Dis. 2013;66(5):428-32. [Link] [DOI:10.7883/yoken.66.428]
11. Alcantar-Curiel D, Tinoco JC, Gayosso C, Carlos A, Daza C, Perez-Prado MC, et al. Nosocomial bacteremia and urinary tract infections caused by extended-spectrum β-lactamase-producing Klebsiella pneumoniae with plasmids carrying both SHV-5 and TLA-1 genes. Clin Infect Dis. 2004;38(8):1067-74. [Link] [DOI:10.1086/382354]
12. Blondeau J, Vaughan D. A review of antimicrobial resistance in Canada. Can J Microbiol. 2000;46(10):867-77. [Link] [DOI:10.1139/w00-076]
13. Teimour Pour M, Gheysarzadeh A, Pakzad I, Valadbeigi H, Maleki A, Sadeghifard N. Antimicrobial resistance and genetic analysis of multi-drug resistant Klebsiella pneumoniae isolates by pulsed-field gel electrophoresis. Gene Rep. 2020;19:100638. [Link] [DOI:10.1016/j.genrep.2020.100638]
14. Azargun R, Sadeghi MR, Soroush Barhaghi MH, Samadi Kafil H, Yeganeh F, Ahangar Oskouee M, et al. The prevalence of plasmid-mediated quinolone resistance and ESBL-production in Enterobacteriaceae isolated from urinary tract infections. Infect Drug Resist. 2018;11:1007-14. [Link] [DOI:10.2147/IDR.S160720]
15. Salah FD, Soubeiga ST, Ouattara AK, Sadji AY, Metuor-Dabire A, Obiri-Yeboah D, et al. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob Resist Infect Control. 2019;8:104. [Link] [DOI:10.1186/s13756-019-0552-0]
16. Ibrahim NH. Frequency of plasmid-mediated quinolone resistance determinants qnr and qepa among clinical isolates of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases from Saudi Arabia intensive care units. Int J Microbiol Res. 2017;9(8):924-9. [Link]
17. Unlu O, Demirci M. Detection of carbapenem-resistant Klebsiella pneumoniae strains harboring carbapenemase, beta-lactamase and quinolone resistance genes in intensive care unit patients. GMS Hyg Infect Control. 2020;15:Doc31. [Link]
18. Nourozi M, Mirkalantari S, Omidi S. Frequency of plasmid-mediated quinolone resistance genes qnrA, qnrB, and qnrS among clinical isolates of Klebsiella pneumoniae. J Appl Biotechnol Rep. 2020;7(4):203-7. [Link]
19. Mohammadbeigi M, Akbarmehr J, Jafari B. Evaluation of the frequency of plasmid-mediated quinolone resistance genes in clinical isolates of Escherichia coli and Klebsiella spp. in Tehran. J Microb World. 2016;9(3):199-207. [Persian] [Link]
20. Ghadami M, Shokoohizadeh L, Mirzaee M. Prevalence of plasmid-mediated quinolones resistance among Klebsiella pneumoniae strains isolated from hospitals in Borujerd, Iran. Med Lab J. 2017;11(3):1-5. [Link]
21. Lavilla S, Gonzalez-Lopez J, Sabate M, Garcia-Fernandez A, Larrosa M, Bartolome R, et al. Prevalence of qnr genes among extended-spectrum β-lactamase-producing enterobacterial isolates in Barcelona, Spain. J Antimicrob Chemother. 2008;61(2):291-5. [Link] [DOI:10.1093/jac/dkm448]
22. Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: A multifaceted threat. Clin Microbiol Rev. 2009;22(4):664-89. [Link] [DOI:10.1128/CMR.00016-09]
23. Liu Y, Du Fl, Xiang TX, Wan LG, Wei Dd, Cao XW, et al. High prevalence of plasmid-mediated quinolone resistance determinants among serotype K1 hypervirulent Klebsiella pneumoniae isolates in China. Microb Drug Resist. 2019;25(5):681-9. [Link] [DOI:10.1089/mdr.2018.0173]
24. Azadpour M, Soleimani Y, Rezaie F, Rezaeifar M. Antibiotic susceptibility pattern and identification of quinolone-resistant strains of Klebsiella pneumoniae clinical specimens from Khorramabad, Iran. Trop Biomed. 2017;34(2):412-8. [Link]
25. Jamshidi MRM, Zandi H, Eftekhar F. Correlation of quinolone-resistance, qnr genes and integron carriage in multidrug-resistant community isolates of Klebsiella spp. Iran J Basic Med Sci. 2019;22(12):1387-91. [Link]
26. Roshani M, Goodarzi A, Hashemi A, Afrasiabi F, Goudarzi H, Arabestani M. Detection of qnrA, qnrB, and qnrS genes in Klebsiella pneumoniae and Escherichia coli isolates from leukemia patients. Rev Res Med Microbiol. 2022;33(1):14-9. [Link] [DOI:10.1097/MRM.0000000000000250]








